The University of Washington (funded-and-in-partnership with Microsoft) have published a new paper on the capabilities of data storage and retrieval using synthetic DNA with a standard base of ATGC. (*this does not include the chances of using synthetic nucleotides such as X/Y or others, an entire topic to get into later).
In a simple proof-of-concept test, the team successfully encoded the word “hello” in snippets of fabricated DNA and converted it back to digital data using a fully automated end-to-end system, which is described in a new paper published March 21 in Nature Scientific Reports.
This is a step towards a data storage method that will ensure that the information we create will be able to sustain itself without intervention for potentially tens of thousands (hundreds of thousands by some accounts) years. The current state of data storage is in a very tight spot with our current strategies - information can quickly (and will) disappear with faulty backups, mismanagement, or antiquated drive systems in the next 20 years.
A more recent example of this would by the loss of geocities where a vast majority of content creation, media, theories, discussions and more of the early internet (1994 - 2009) have become impossible to view or see as the systems in place have gone away for eternity. Another recent example would be Myspace which just in the last few weeks confirmed that their entire musical archives from 2005-2015 were corrupted in a "data migration" and now without any way of retrieval. Over a decade of music on what was one of the first musical social networks are now gone as if it never existed in the first place.
"If a tree falls in a forest and no one is around to hear it, does it make a sound?" In the case of Myspace - what tree?
There are several reasons for this "Digital Dark Age" where most of which will be due to server costs (hardware/bandwidth), personnel costs (management/upkeep), and pure negligence. There are several organizations working towards the storage of all information created on the internet - Internet Archive (Wayback Machine), Archive.is, and many others. This will not stop the digital dark age from happening, as already mentioned geocities, myspace, and millions of other communities have disappeared into the aether and became one with the void, but at the very least someone is trying to save out history.
Now, a proposed method that would stop the future destruction of our modern reality is by embracing DNA data storage systems.
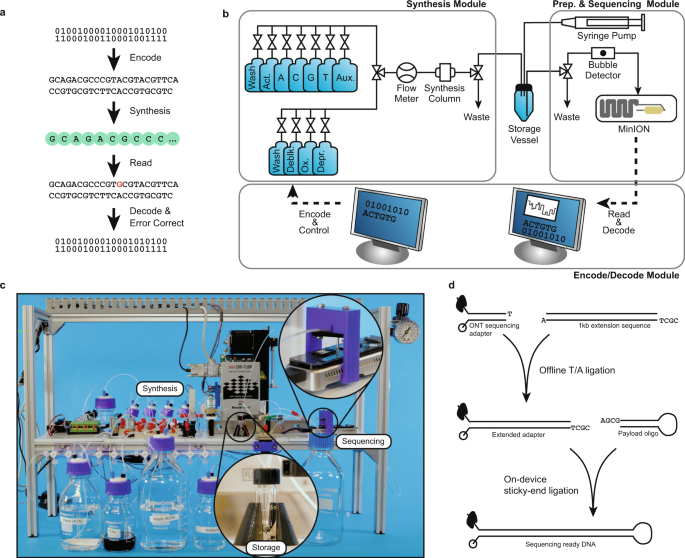
How it works
"Information is stored in synthetic DNA molecules created in a lab, not DNA from humans or other living things, and can be encrypted before it is sent to the system. While sophisticated machines such as synthesizers and sequencers already perform key parts of the process, many of the intermediate steps until now have required manual labor in the research lab. But that wouldn’t be viable in a commercial setting", said Chris Takahashi, senior research scientist at the UW’s Paul G. Allen School of Computer Science & Engineering “You can’t have a bunch of people running around a datacenter with pipettes — it’s too prone to human error, it’s too costly and the footprint would be too large,”.
But, The UW team, in collaboration with Microsoft, are developing a programmable system that automates lab experiments by harnessing the properties of electricity and water to move droplets around on a grid of electrodes. The full stack of software and hardware, nicknamed “Puddle” and “PurpleDrop,” can mix, separate, heat or cool different liquids and run lab protocols.
The automated DNA data storage system uses software developed by the Microsoft and UW team that converts the ones and zeros of digital data into the As, Ts, Cs and Gs that make up the building blocks of DNA. Then it uses inexpensive, largely off-the-shelf lab equipment to flow the necessary liquids and chemicals into a synthesizer that builds manufactured snippets of DNA and to push them into a storage vessel.
When the system needs to retrieve the information, it adds other chemicals to properly prepare the DNA and uses microfluidic pumps to push the liquids into other parts of the system that “read” the DNA sequences and convert it back to information that a computer can understand.
The cost of production
The current cost of the UW/Microsoft system is roughly ten thousand dollars using off the shelf mechanical hardware and software. While the current rate of "Hello" per 21 hours is not ideal - the UW research team believes they can achieve far faster results in a very near future by cutting it down to just a few hours. While this is not ideal for mass production and usage- as a proof-of-concept this is a fundamental game changer for our data-centric modern reality, and our increasingly data-centric future.
There is a lot of research in this area and this is just the start, I am looking forward to our future data architectures be built on the same core structures as all of us, DNA, a more cost effective, smaller, data system.
